


Research
We use genetic and genomic tools to shed light on behavioral and evolutionary questions. Our projects primarily focus on populations for which we can combine genetic analysis with data on the behavior, life history, and environmental milieu of known individuals. This approach allows us to connect behavioral and environmental effects on trait variation in individuals, to evolutionary outcomes on the level of populations and species.
It also gives us the opportunity to study the role of gene regulation in explaining social environmental effects on fertility, health, and survival across the life course, in mammalian models for human health.
Much of the research in the lab focuses on a population of baboons in the Amboseli ecosystem of Kenya, which has been the subject of research by the Amboseli Baboon Research Project for almost 50 years. Check out the ABRP's webpage and Facebook page! We also study rhesus macaques in captivity and, through a collaboration with the Kalahari Meerkat Project, meerkats and Damaraland mole rats in South Africa.
Current projects

Social and environmental adversity can have long-lasting impacts on both fitness and health. For instance, low social status and social isolation in early life have been linked to elevated rates of cardiovascular disease later in life, even when status differences are erased. At the same time, traits that are influenced by social conditions early in life remain responsive to later life environmental conditions. Epigenetic mechanisms have been hypothesized to mediate these effects, but little is known about social, behavioral, and environmental epigenetics in natural populations.
We are taking advantage of the extensive longitudinal data on the Amboseli baboon population--including information on social relationships, social status, resource availability, and life history--to investigate the epigenetic consequences of naturally occurring environmental variation. We are interested in the timing, magnitude, and reversibility of epigenetic effects; the locations in the genome that are most sensitive to environmental conditions; and the impact of epigenetic changes on downstream phenotypes. The results of our research will shed light on the role of epigenetics in linking environmental conditions to fitness-related traits, including health-related outcomes.
This work is a collaboration with Xiang Zhou (Michigan). It is supported by NIH R01HD088558 and F32-HD095616, with past support from NIH R21AG049935-01 and NSF BCS-1455808.

Socially induced stress is strongly correlated with disease susceptibility and mortality risk in humans, and recent studies show similar patterns in nonhuman primates. Rhesus macaques in captivity present the opportunity to study social environmental effects on gene regulation in a controlled setting, and potentially to establish causality. We take advantage of the ability to manipulate social status and social group composition to test the effects of these variables on gene regulation and other interesting phenotypes.
Our studies are revealing the role of different cell types in the response to social status-mediated stress, the plasticity of these effects when social status changes, and the degree to which social status affects an animal's ability to respond to other environmental challenges. This work is a collaboration with Luis Barreiro (University of Montreal) and Vasiliki Michopoulos (Emory) and has been funded through NIH grants 5R01GM102562 and R01AG057235.
We also complement our work in captive primates with studies in the Amboseli baboons. There, we test whether demographic characteristics and social history predict variation in gene regulation, particularly in the immune system. Our studies show that social status is a key predictor of gene regulation in male baboons, but much less so in females, and may indicate overall condition. This work has been funded in part through NSF BCS-1455808, the Leakey Foundation, and the Triangle Center for Evolutionary Medicine.

Integrating genomic approaches into studies of natural populations is an exciting emerging area of research. However, vertebrate non-model systems, especially large mammals, face challenges that differ from those presented by studies of traditional model organisms. Motivated by our own experiences, we develop new methods, both lab and computational, for moving the field forward. We have developed:
*Capture protocols to perform whole genome re-sequencing from non-invasively collected samples
*Software to perform pedigree reconstruction and local ancestry estimation from low coverage resequencing data [WHODAD, with Sayan Mukherjee (Duke) and Xiang Zhou (Michigan); LCLAE, with Jeff Wall (UCSF)]
*Methods to model bisulfite sequencing data in populations that contain kin or complex population structure [MACAU, with Xiang Zhou], and a user-friendly R Shiny app for accompanying power analysis
*Functional genomic approaches to test the causal effects of variation in DNA methylation on gene expression [mSTARR-seq, with Tim Reddy (Duke) and Luis Barreiro (Montreal)]
All of our software and protocols are freely available, as are select reagents. For more details, see the Protocols and Software section of the website. Components of this work have been supported by NSF DEB-1405308, NIH R21AG049935-01, NIH 5R01GM102562, NSF BCS-1455808, and a Sloan Early Career Research Fellowship.

Hybridization and admixture have been deeply important in primate evolutionary history. The Amboseli baboon population is located in an active hybrid zone: while the dominant baboon species in Amboseli is the yellow baboon (Papio cynocephalus), the ABRP has observed periodic immigration of anubis (P. anubis) or hybrid baboons into the basin since 1982. From this vantage point, we have the rare opportunity to investigate both the causes and consequences of admixture between two closely related primates.
Our work has revealed that anubis ancestry is associated with earlier maturation and a mating advantage to males, and that the vast majority of Amboseli animals are admixed to some degree. Despite no obvious phenotypic costs ot admixture, however, the hybrid zone appears to be narrow, transitioning quickly between phenotypically anubis animals and phenotypically yellow animals. These observations present a puzzle that we aim to solve, using a combination of behavioral, demographic, and genomic data.
We pursue this line of research collaboratively with Susan Alberts (Duke University) and Jeff Wall (UCSF), with support from NSF BCS-1751783.
Photo Credit: Noah Snyder-Mackler
A freezer of noninvasively collected baboon samples
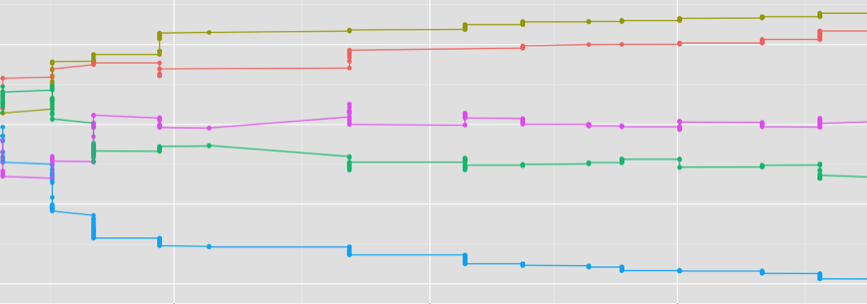
Attainment and stability of macaque dominance ranks over time (each line shows Elo-based rank for an individual female)
An infant baboon in Amboseli rides jockey style on her mother's back
With Elizabeth Archie (University of Notre Dame), Ran Blekhman (University of Minnesota), Luis Barreiro (University of Montreal), and the Earth Microbiome Project, we are investigating the signature of social structure and social interactions on the gut microbiome in the Amboseli baboons (supported in part through NIH/NICHD 2P2C-HD065563-06).
With Carol Shively (Wake Forest University), we are investigating how diet and social status combine to influence the gut microbiome in captive long-tailed macaques.
Using data from Amboseli, we are investigating the factors that predict, and are predicted by, epigenetic aging in wild baboons (supported in part through Duke's Center for Population Health and Aging), and using data from Amboseli and other baboon species, we are performing comparative analyses of DNA methylation evolution (supported by NSF BCS-1751783)
Baboons on the savanna, Amboseli, Kenya
Competition is a central tenet of Darwinian evolution. In contrast, cooperative behavior poses a challenge to evolutionary thought, with special resonance because of the high levels of cooperation in humans. In the most cooperative animal societies, even production of surviving offspring requires cooperative effort, and members of a social group often adopt alternative social roles (e.g., as breeders or helpers). However, outside eusocial insects, we know little about the molecular changes or trade-offs that characterize transitions between roles.
We are combining behavioral, genomic, and shape analysis methods to study these transitions in two of the most cooperative nonhuman mammals yet described, the meerkat and the Damaraland mole rat. In the meerkats, we will test the hypothesis that the competing demands of growth, immune defense, and reproduction are observable at the transcriptional level, and that these demands are resolved differently by helpers and breeders. In the Damaraland mole rats, we are studying how experimentally induced transitions to queen status influence the physiology and genomics of skeletal growth and aging.
The meerkat work takes advantage of a three-decade study of wild meerkats at the Kalahari Meerkat Project in South Africa, and is a collaboration with Tim Clutton-Brock (Cambridge University), Sayan Mukherjee (Duke University), and Luis Barreiro (University of Montreal). It is supported by the Human Frontiers Science Program RGP0051. The Damaraland mole rat work is a collaboration with Tim Clutton-Brock and is supported by NSF IOS-1749419 and the European Research Council.
Photo Credit: Jenny Tung
Wild meerkats at the Kalahari Meerkat Project, South Africa
The impact of social adversity on gene regulation
Epigenetic consequences of environmental variation across the life course
Gene regulation in cooperative societies
Methods development for genomics in natural populations
The causes and consequences of hybridization in baboons
Anubis baboons, Samburu, Kenya
Yellow baboons, Tsavo, Kenya
Other projects in the lab